Both the Daylight Theory Manual and OpenSMILES Specification explain the concept for the organic subset. The specifications provide a more detailed explanation but as a simple example with the SMILES '[CH2]=[CH][CH3]' we can omit hydrogens counts and just write 'C=CC'. However, with '[CH]=[CH][CH3]' we must include the count on the first atom '[CH]=CC'. We include the labelled count if the implied count does not match.
The OpenSMILES specification explains how we can determine the implied hydrogen count.
"The implicit hydrogen count is determined by summing the bond orders of the bonds connected to the atom. If that sum is equal to a known valence for the element or is greater than any known valence then the implicit hydrogen count is 0. Otherwise the implicit hydrogen count is the difference between that sum and the next highest known valence." - OpenSMILES
This approach is simple and elegant for the aliphatic subset with non-aromatic bonds but what about the aromatic subset? One could speculate we have to assign explicit bond orders (kekulizé) a molecule to obtain the correct counts. Indeed the OpenSMILES specification seems to suggest that.
"...the SMILES software must verify that electrons can be assigned without violating the valence rules..."
Personally, I do not believe this to be the case and having the correct hydrogen count provides an additional validation if we do assign a kekulé structure.
Perhaps how we get the count for the aromatic subset is obvious and doesn't need including in the specification. However as I will show there are a couple approaches used by the different (open source) toolkits.
Disclaimer: I may be completely wrong but given the lack of documented explanation on the issue I believe it's important to at least collect some ideas on how it is currently done and how it could be done.
Parsing in Toolkits
It appears that many toolkits use their own internal valence model rather than one specific to SMILES. It is difficult to know all the intricacies of each toolkit but here's what I see from a high level view. Any maintainers of the respective toolkits please let me know of any mistakes and I'll add a correction.The CDK uses it's own valence model via a decision tree based atom typer. The atom typer assigns a formal neighbour count which is then used to infer the number of hydrogens. This piece of code is from the end of the current SmilesParser.java:258-276. The decision tree in CDK is a large if else cascade checking possibly exceptions.
Code 1 - CDK atom typing
// CDK org/openscience/cdk/smiles/SmilesParser.java lines 258-276. IAtomType type = matcher.findMatchingAtomType(molecule, atom); boolean isAromatic = atom.getFlag(CDKConstants.ISAROMATIC); AtomTypeManipulator.configure(atom, type); atom.setFlag(CDKConstants.ISAROMATIC, isAromatic);
Code 2 - OpenBabel atom typing
// Open Babel src/formats/smileyformat.cpp lines 278-288 OBAtomTyper typer; typer.AssignImplicitValence(*pmol);
The other parser assigns bond orders to aromatic bonds after parsing. The code snippet below is at the end of the parser (smilesformat.cpp:516-520). With set bond orders the counting is the same as with the aliphatic subset.
Code 3 - OpenBabel atom typing additionals
// Open Babel src/formats/smilesformat.cpp lines 516-520
//set aromatic bond orders
mol.SetAromaticPerceived();
FindAromaticBonds(mol);
FindOrphanAromaticAtoms(mol);// CM 18 Sept 2003
mol.AssignSpinMultiplicity();
mol.UnsetAromaticPerceived();
RDKit again uses it's own internal valence model. This model is invoked when parsing SMILES if sanitise is set (SmilesParse.cpp:184-198). Interestingly the RDKit internal model uses a similar approach to Open Babel and my first implementation. Frowns also uses the same approach and we will come back to this later.
Code 4 - RDKit Sanitize
// RDKit Code/GraphMol/SmilesParse/SmilesParse.cpp lines 184-198
if(sanitize && res){
// we're going to remove explicit Hs from the graph,
// this triggers a sanitization, so we do not need to
// worry about doing one here:
try {
ROMol *tmp = MolOps::removeHs(*res,false,false);
delete res;
res = static_cast<RWMol *>(tmp);
// figure out stereochemistry:
MolOps::assignStereochemistry(*res,true);
} catch (...) {
delete res;
throw;
}
}
The recent parser written in Scala, chemf, uses a concise decision tree of what aromatic combinations "make sense" (SmilesAtom.scala:38).
Code 5 - Chemf
/* Snippet from chemf chemf.parser.SmilesAtom.scala lines 45 - 62*/
bs count (Aromatic ==) match {
case 1 ⇒ default (2 + (bs foldMap (_.valence)))
case 0 ⇒ default (bs foldMap (_.valence))
case _ ⇒ bs.toList sortBy (_.valence) match {
case Aromatic::Aromatic::Aromatic::Nil ⇒
if (e == C || e == N || e == P || e == B) 0.success else fail
case Aromatic::Aromatic::Single::Nil ⇒
if (e == C || e == N || e == P || e == B) 0.success else fail
case Aromatic::Aromatic::Double::Nil ⇒
if (e == C || e == S) 0.success else fail
case Aromatic::Aromatic::Nil ⇒
if (e == C || e == B) 1.success
else if (e == N || e == P || e == O || e == S) 0.success
else fail
//other combos with 2 or more aromatic bonds don't make sense
case _ ⇒ fail
}
}
Code 6 - OUCH
-- snippet from Ouch/Structure/Molecule.hs lines 380-391
-- n neighbors is larger than the first valence + second
-- valence => identity (do nothing)
outputH | nb >= ((fst val) + (abs(snd val))) = m
-- There is one or more bonds to a radical and the atom
-- is aromatic => recurse with a molecule that has electrons added (mEl)
| nbrB && Set.member AromaticAtom (atomMarkerSet a) = fillValenceAtIndex mEL i
-- Fill empty valences with hydrogen => recurse with a molecule that has
-- an extra hydrogen on this atom index (mH)
| nba < fst val = fillValenceAtIndex mH i
-- Then, fill lone-pairs if needed => recruse with a molecule that has an
-- extra lone pair on this atom index (mLp)
| nb < ((fst val) + (abs(snd val))) = fillValenceAtIndex mLP i
-- Pattern completion (default) => identity
| otherwise = m
In the above function the bond order sum is used. Interestingly aromatic bonds contribute '2' to the sum (Atom.hs:136-148).
Code 7 - OUCH snippet 2
-- Snippet from Ouch Ouch/Structure/Atom.hs lines 136-148
occupiedValence :: Atom -> Integer
occupiedValence a = output
where output = Set.fold (\b acc -> acc + bondOrder b) 0 (atomBondSet a)
bondOrder b = case b of
Sigma {} -> 1
Pi {} -> 2
PiPi {} -> 3
Aromatic {} -> 2
Delta {} -> 4
Hbond {} -> 0
Ionic {} -> 0
Antibond {} -> 1
Any {} -> 0
PerlMol has a valence model specific to SMILES. The parser doesn't have the aromatic bond token ':' and uses a bond order of '1' for an implicit bond. The hydrogen count is computed as per the aliphatic subset - if the atom is an aromatic carbon or nitrogen the hydrogen count is decreased.
Code 8 - PerlMol
# Snippet from PerlMol Chemistry/File/SMILES.pm lines 412-423
# returns the number of hydrogens for an atom, assuming it has
# no charge or radical (because those require an explicit H-count anyway)
sub calc_implicit_hydrogens {
my ($self, $atom) = @_;
no warnings 'uninitialized';
my $h_count = $ORGANIC_ELEMS{$atom->symbol} - $atom->valence;
if ($atom->attr("smiles/aromatic") and $atom->symbol =~ /^[CN]$/) {
$h_count--;
}
$h_count = 0 if $h_count < 0;
$h_count;
}
It's clear there are different approaches and it's tempting to speculate there are still some more differences in closed source toolkits.
Generation
Of course the implicit hydrogen count of the aromatic subset also maters when generating SMILES. One well known example is of aromatic nitrogens which contribute a lone pair - for example pyrrole.| 1H-pyrrole |
The CDK, Open Babel and RDKit handle this as a special exception when generating the SMILES string. The CDK code refers to this as a special nitrogen (SmilesGenerator.java:1652-1657).
Code 9 - CDK's special nitrogen
// we put in a special check for N.planar3 cases such
// as for indole and pyrrole, which require an explicit
// H on the nitrogen. However this only makes sense when
// the connectivity is not 3 - so for a case such as n1ncn(c1)CC
// the PLANAR3 N already has 3 bonds, so don't add a H for this case
boolean isSpecialNitrogen =
a.getSymbol().equals("N") &&
a.getHybridization() == IAtomType.Hybridization.PLANAR3 &&
container.getConnectedAtomsList(a).size() != 3 &&
(a.getFormalCharge() == null || a.getFormalCharge() == 0);
brackets = brackets | isSpecialNitrogen;
Similar to CDK, Open Babel has the method CorrectAromaticAmineCharge (smilesformat.cpp:2569-2592)
Code 10 - OpenBabel CorrectAromaticAmineCharge
void OBMol2Cansmi::CorrectAromaticAmineCharge(OBMol &mol)
{
OBAtom *atom;
vector::iterator i;
_aromNH.clear();
_aromNH.resize(mol.NumAtoms()+1);
for (atom = mol.BeginAtom(i);atom;atom = mol.NextAtom(i))
if (atom->IsNitrogen() && atom->IsAromatic())
if (atom->GetHvyValence() == 2)
{
if (atom->GetValence() == 3 || atom->GetImplicitValence() == 3)
_aromNH[atom->GetIdx()] = true;
}
}
These hardcoded exceptions work, unless you miss an exception...
| 1H-phosphole |
Neither CDK or the default Open Babel parser will include the required hydrogen. Note - obabel v 2.3.1 (latest on Mac), this might have been fixed in v 2.3.2..
Code 11 - OpenBabel aromatic phosphorus
[intrepid ~]: obabel -:"[nH]1cccc1" -osmi [nH]1cccc1 1 molecule converted [intrepid ~]: obabel -:"[pH]1cccc1" -osmi P1CCCC1 1 molecule converted
Adding another conditional (as in RDKit) for aromatic phosphate solves this problem (SmilesWirte.cpp:75-69).
Code 12 - RDKit non-standard handling
// another type of "nonstandard" valence is an aromatic N or P with
// explicit Hs indicated:
if((num==7||num==15) && atom->getIsAromatic() && atom->getNumExplicitHs()){
nonStandard=true;
}
Although this will handle most (possibly all) cases if there is a way to assign implicit hydrogen counts no special treatment is needed. As with the aliphatic subset if the implicit hydrogen count doesn't match what we have stored we must include the hydrogen counts.
Fractional Bond Orders
As mentioned earlier, my first approach was the same as OpenBabel, RDKit's and Frowns (something I didn't realise until collecting the information for this post). The approach seems obvious - aromatic bonds have bond order '1.5'. This is of course not true but we will see how this model fits examples.To fix the problematic examples we need an additional rule. Only check the default valence for aromatic atoms. A justification for this could be that elements which have more than one valence cannot go to a higher valence state as the electrons which would bond those hydrogens are contributed to the pi bonding system. We can see this is also the approach used by RDKit - although RDKit doesn't allow 5 valent nitrogen so this particular example won't occur. Also note that the partial bond order sum is rounded up.
Code 12 - RDKit valence calculation
/* RDKit/Code/GraphMol/Atom.cpp lines 193-232*/
// check accum is greater than the default valence
unsigned int dv = PeriodicTable::getTable()->getDefaultValence(d_atomicNum);
int chr = getFormalCharge();
if(isEarlyAtom(d_atomicNum)) chr*=-1; // <- the usual correction for early atoms
if (accum > (dv + chr) && this->getIsAromatic()){
// this needs some explanation : if the atom is aromatic and
// accum > (dv + chr) we assume that no hydrogen can be added
// to this atom. We set x = (v + chr) such that x is the
// closest possible integer to "accum" but less than
// "accum".
//
// "v" here is one of the allowed valences. For example:
// sulfur here : O=c1ccs(=O)cc1
// nitrogen here : c1cccn1C
int pval = dv + chr;
const INT_VECT &valens = PeriodicTable::getTable()->getValenceList(d_atomicNum);
for (INT_VECT_CI vi = valens.begin(); vi != valens.end() && *vi!=-1; ++vi) {
int val = (*vi) + chr;
if (val > accum) {
break;
} else {
pval = val;
}
}
accum = pval;
}
// despite promising to not to blame it on him - this a trick Greg
// came up with: if we have a bond order sum of x.5 (i.e. 1.5, 2.5
// etc) we would like it to round to the higher integer value --
// 2.5 to 3 instead of 2 -- so we will add 0.1 to accum.
// this plays a role in the number of hydrogen that are implicitly
// added. This will only happen when the accum is a non-integer
// value and less than the default valence (otherwise the above if
// statement should have caught it). An example of where this can
// happen is the following smiles:
// C1ccccC1
// Daylight accepts this smiles and we should be able to Kekulize
// correctly.
accum += 0.1;
A similar approach is used in Frowns library. The snippet below shows the use of a 1.5 bond order from Atom.py:82-92.
Code 14 - FROWNS sumBondOrders
def sumBondOrders(self):
result = 0
for x in self.bonds:
if x.bondtype == 4:
# XXX FIX ME
# this is a hack to fix bad conjugation
# this will be fixed soon
result += 1.5
else:
result += x.bondorder
return result
Open Babel also uses a bond order of '1.5'. Although in their case they actually count electrons and then divided by 2 (atom.cpp:1000-1015). This avoids fractional bond orders - at least until the division where the fractional part is truncated (5/2 = 2).
Code 15 - OpenBabel valence
// Open Babel src/atom.cpp lines 1000-1015
for (bond = ((OBAtom*)this)->BeginBond(i);bond;bond = ((OBAtom*)this)->NextBond(i))
{
bo = bond->GetBO();
bosum += (bo < 5) ? 2*bo : 3;
}
bosum /= 2;
return(bosum);
Using this model will work very well and give use correct results. There are however still some questions, in particular the ambiguity and having to round bond order sums is troublesome.
Also we need to know which bonds are aromatic - 'cc' is an aromatic bond and 'CC' is a single bond if we have these as an implicit bond as suggested by SMILES we need to check the atoms in each case to determine if the order is '1' or '1.5'. Additionally I found a couple of examples in ChEMBL where up ('/') and down ('\') bonds are part of an aromatic system (contrived example below). Again we need to check the atoms of those bonds to see if they are aromatic or single. These problems only occur if we have bond labels specifically for handling SMILES but it would be nice to have something more elegant.
 |
| C\C=c1/ccc(=C)cn1 |
Atom-based
After using the fractional bond order and testing on some datasets I was relatively happy it solved the problem. Using a bond order of 1.5 will certainly work - pragmatic, yes - elegant, no.Despite this I couldn't shake the feeling that "Dijkstra would not have liked this". I think this is also evident from the comments in the RDKit and Frowns source code (see above).
What I now believe is the correct way, or at least closer to the Daylight way is that instead of modifying the bond orders we modify the starting value of our bond order sum. With aromatic atoms, instead of starting at 0 we start at 1.
The point of the 1.5 bond order is to increase the bond order sum to be higher for aromatic atoms. We can achieve the same thing by simply changing the starting value of the sum. This also fits in that aromaticity in SMILES is primarily defined by atoms and not bonds. There is the ':' aromatic bond but it is not required (and rarely/never used).
In practice this means implicit, single, aromatic, up and down bonds now all contribute 1 to the bond order sum. Of course this is completely bogus but we're not saying that aromatic bond is the same as a single bond - rather just that it contributes a bond order of '1'. This also means that the implicit bond between aliphatic and aromatic atoms 'cc', 'CC' and 'cC' would all contribute the same to the bond order and we can treat them the same.
This is particularly useful for cases like '[Te]1cccc1' (ChemSpider:119908). Using a bond order of 1.5 we would determine the bond order sum of each carbon adjacent to the '[Te]' to be '2.5' or '2' (5/2). Rounding up or down we get '1' (correct) or '2' hydrogens.
It would seem that PerlMol was nearly there - the implicit bond order in PerlMol is '1' and the hydrogen count was decreased for aromatic atoms. This was only done for carbon and nitrogen though and should be applied to all aromatic atoms.
Is it Correct?
I can not say for certain whether this model is correct but it is certainly closer than using a fractional bond order.The well known Depict service from Daylight displays the structure diagram for a given SMILES string. It will also report problems with SMILES. For example it reports that [C]C has unusual valence 1 (normal 2).
Now what is really interesting is if we give it some aromatic atoms. It turns out that a single aromatic atom [c] also has unusual valence 1 (normal 2). But wait - there aren't any bonds to that atom but it has valence of 1?
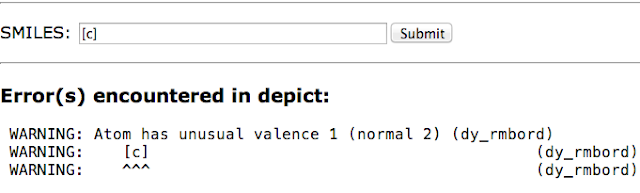 |
| Aromatic carbon has no neighbours but has a valence of 1? |
This also explains why c is '[CH3]'.
Using this approach also allows us to get the correct count for some strange examples. One example could be an acyclic aromatic atom (rejected by some toolkits). This can happen for example with an exocyclic oxygen (oc1ccocc1). Using this approach we reach the correct answer that there are no hydrogens on that sprouting oxygen. This can then be used in the assignment of a correct Kekulé structure. One can see how this SMILES string is interpreted 4 different ways by various toolkits - oc1ccocc1 (Ambit).
With this model it also means that when we generate the SMILES we can now check how many implicit hydrogens aromatic atoms should have. Doing so we would say that for pyrrole the implied hydrogen count would be '0'. This does not match our stored value and so we need to include the bracket atom with the labelled hydrogen count.
Complications
There are of course still some complications - which seem to be when we mix aromatic and aliphatic parts. The fact that the first case here is read as cyclohexane further shows there isn't an aromatic bond order used.1. C1:C:C:C:C:C1 (daylight reads as cyclohexane)
2. C=1:C=C:C=C:C1
3. c=1c=cc=cc1 (problematic)
4. c-1c-cc-cc1
With the described model we can't reach the correct answer for (3.). You're unlikely to encounter such a case in a clean dataset but we can modify our model slightly to get the correct results.
Instead of starting at '1' we add '1' to the bond order sum of aromatic atoms if the bond order sum is the same as the number of explicit neighbours. Indeed this agrees with Depict that 'c[c]cc' has a valance of 3 but '[c]=c' has a valence of 2 (if we started at '1' it would also be '3').
Hopefully this has provided some insight or at least show how the implicit hydrogens can be counted in aromatic SMILES.
Supplementary
Re-reading back through the mailing list it seems this has of course be discussed (at least partially) - no decision was made. As usual the sourceforge mailing archives are a pain to find stuff in but the discussion [1] is summarised on the OpenSMILES repository [2].
As I see it there are three models which can be used.
- Use bond order of 1.5 for aromatic bonds.
- Reduce the hydrogen count for aromatic atoms - also attempts to allow this to specify radicals (C1CCcCC1). This has many problems but that would need another post.
- Increase the valence by 1 if there are no double/triple bonds.
Mailing List Links
- http://sourceforge.net/mailarchive/forum.php?thread_name=60825b0f0709302037g2d68f2eamdb5ebecf3baea6d1%40mail.gmail.com&forum_name=blueobelisk-smiles
- https://github.com/timvdm/OpenSMILES/blob/master/discussion_summary.txt
- OpenSMILES and aromaticity - Andrew Dalke
- Smiles and Aromaticity: Broken? - Rich Apodaca





No comments:
Post a Comment
Note: only a member of this blog may post a comment.